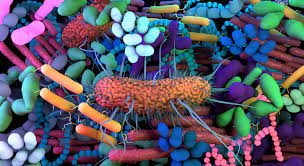
This working session will take place from February 7 - February 14, 2025
This year, we will bring together ~50 researchers and trainees to discuss new methods and cutting-edge approaches for high-throughput sequencing analysis. The theme is virus and microbial genomics Technologies to profile these extremely complex communities of ‘’small organisms’’ have developed very quickly and it is now routine and cost effective to sample the microbiome across a broad range of domains.
The days are organized as discussions around specific themes, with one-on-one exchanges between experts and trainees. The invitees this year cover domains such as the microbiome of the human gut/oral/respiratory/urogenital in the context of health and disease; clinical metagenomics; of animals in relation to food production systems; the microbiome and diet; fungal viruses; approaches for identifying new microbes. The meeting will also cover high-throughput systems level approaches for studying microbes including genetic screens, single-cell bacterial/fungal profiling, microfluidic sample handling, improved CRISPR/CasX modification toolkits, synthetic biology along with core biostatistical computational approaches including generative modeling in microscopy, sample decomposition and compositional data (CODA) analyses.
Computational Paradigms in Molecular Biology Revisited will take place from February 2 - February 9, 2024
The past decade has witnessed the rapid development of new computational techniques and paradigms in computational biology. These approaches emerged from the machine learning, probabilistic modeling, statistical inference and artificial intelligence communities and have been further refined within the context of a broad range of applications in molecular biology. This meeting brings together researchers who have expertise with generative modeling and deep learning across a range of different applications including structural biology, drug repositioning and discovery, generative models for single cell analyses, metagenomics, and molecular imaging. Many of our invitees are also committed to the development of computational devices with clinical utility. Such endpoints also raise important ethical and socioeconomic considerations that also must be addressed for AI-related tools to be fully incorporated into modern molecular biology and clinical applications.
Cells in Space will take place from January 27th - February 3rd, 2023.
This year will be our 21st Systems Biology workshop: over the past two decades, these meetings have brought together individuals working in the diverse range of fields.
This meeting brings together researchers interested in exploring cells in their context. Recent advances have reinforced the importance of understanding where cells are localized in tissues to further characterize their similarity and differences (cell types) and shed light on their interactions, communication and functionality.
The invitees collectively champion the development and/or application of EXPERIMENTAL approaches to identify the physical location of cells based on sequencing or imaging methods, and COMPUTATIONAL models to analyze cellular information and/or predict its location.
We are excited to extend discussions to how a wide variety of external factors and stressors, such as space travel, the exposure to microgravity and space radiation, the synergistic effect of microbiomes, infection processes, tumor microenvironments, etc., influence or alter cellular expression and epigenetics programs.
Encoding an Decoding Function in the Genome. Friday February 4th to Friday February 11th, 2022
 |
This meeting brings together researchers interested in approaches that systematically modify and perturb genomes in order
- [DECODE] to understand the function of the target loci in terms of gene expression and regulation;
- [ENCODE] to modify the function of the target loci to change its expression and regulation for specific goals.
The invitees collectively champion modern -omic approaches that often include precise genome editing (eg CRISPR/Cas9) applied to populations in a high-throughput manner using different strategies (eg lentiviral infections or robotic delivery), tracked in different ways (eg competitive growth assays versus single cell droplet encapsulation), and profiled with -omic and microscopy technologies. Such systematic screens are central to exploring
- to build codebooks describing interactions between DNA, RNA, protein, and genome struture;
- to decipher the multivariate biology of higher order genetic and biochemical interactions;
- to understand the interconnected relationships in systems from ecology, health, and disease;
- to efficiently optimize systems in the context of synthetic biology.
Emerging Model Systems. January 24th-31st.
 |
This meeting brings together researchers interested in the development of new model organisms. Such models either facilitate exciting new questions that can be asked for the first time, or facilitate unprecedented ability to explore long-standing fundamental questions central to the life sciences. The emergence of many new “non-model” model systems recently is at least in part due to recent advancements in gene editing technologies, cellular imaging, and single cell genomics. Therefore, the workshop also seeks to bring investigators interested in the advancement of such techniques to better exploit the unique biological features of these organisms.
The invitees collectively champion emerging model systems across a broad swath of the Tree of Life. This includes
- Protist, bacterial and fungal unicellular model systems for studying, for example, carbon sequestration in marine sytems, single cellular regeneration, and genome organization.
- Individuals who have developed novel models in multicellular organisms including sytems to study regeneration, tissue morphology, and neural function.
- Researchers with models that may more accurately recapitulate key aspects of human disease in comparison to current systems.
- Researchers interested in the development of models to study organismal interactions in different environments.
- Experts in developing synthetic biology, -omics, microscopy and computational/statistical biology approaches to model systems.
SC-SEQ-&-MPRAs
This meeting brings together researchers interested in the development of single cell (SC-Seq) sequencing-based approaches and massively parallel reporter assays (MPRAs), in addition to computational bioogists interested in asking new types of questions facilitated by these technologies in variety of biological systems.
 |
Unless indicated otherwise, the talks are scheduled to be 30 minutes.
Here we focused on the genomic of coral and its microbiome.
 |
| Organizers | Institution |
| David Walsh | Concordia University, Canada |
| Vanessa Dumeaux | Concordia University, Canada |
| Mike Hallett | Concordia University, Canada |
| Arthi Ramachandran | Concordia University, Canada |
| Shawn Simpson | Concordia University, Canada |
| Kaitlin Laverty | University of Toronto, Canada |
| Invitees | Institution |
| Amy Apprill | Woods Hole Oceanographic Institute |
| John Archibald | Dalhousie University, Canada |
| Debashish Bhattacharya | Rutgers University |
| Rob Beiko | Dalhousie University, Canada |
| Stefan Bertilsson | Uppsala University, SciLifeLabs |
| Erin Bertrand | Dalhousie University, Canada |
| Adrienne Correa | Rice University |
| Ford Doolittle | Dalhousie University |
| Stephen Giovannoni | Oregon State University |
| Hans-Peter Grossart | Leibniz-Institute of Freshwater Ecology and Inland Fisheries |
| Lee Henry | Queen Mary, University of London |
| Tim Hughes | University of Toronto, Canada |
| Paul Jensen | Scripps, UCSD |
| Andrew Lang | Memorial University, Canada |
| Julie Laroche | Dalhousie University, Canada |
| Connie Lovejoy | Universite Laval, Canada |
| Marie-Jean Meurs | UQAM, Canada |
| Monica Medina | Pennsylvania State University |
| Adam Monier | University of Exeter |
| Quaid Morris | University of Toronto, Canada |
| Spencer Nyholm | University of Conneticut |
| Martin Polz | MIT |
| Francisco Rodriguez Valera | Universitas Miguel Hernandez |
| Andrew Roger | Dalhousie Univeristy |
| Forest Rohwer | San Diego State University |
| Daniel Sher | Haifa University |
| Derek Skillings | University of Bordeaux/CNRS |
| Uli Stingl | University of Florida |
| Shinichi Sunagawa | ETH Zurich |
| Adrian Tsang | Concordia University |
| Doug Wallace | Dalhousie University |
| Harm van Bakel | Mount Sinai |
| Richard Bonneau | NYU |
| David Gresham | NYU |
Here we focused on the role of the immune system in cancer progression, and on the development of cancer immunotherapies.
 |
The focus of this meeting had much of the CCBR-Donnelly in attendance. It was on genetic interaction networks of different types in model organisms.
 |
| Organizers | Institution | |
| Brenda Andrews | University of Toronto | |
| Vanessa Dumeaux | McGill University | |
| Mike Hallett | McGill University | |
| Organizing committee | ||
| Ben Grys | University of Toronto | |
| Natasha Pascoe | University of Toronto | |
| Dara Lo | University of Toronto | |
| Michael Costanzo | University of Toronto | |
| Invitees | ||
| Fritz Roth | University of Toronto | |
| Charlie Boone | University of Toronto | |
| Andy Fraser | University of Toronto | |
| Brendan Frey | University of Toronto | |
| Marian Walhout | University of Massachusetts | |
| Malcolm Whiteway | Concordia University | |
| Tim Hughes | University of Toronto | |
| Alejandro Reyes | EMBL, Germany | |
| Olga Troyanskaya | Princeton University | |
| Steve Scherer | University of Toronto | |
| Mike Tyers | IRIC, University of Montreal | |
| Maitreya Dunham | University of Washington | |
| Chris Loewen | University of British Columbia | |
| Dave Gresham | NYU | |
| Doug Fowler | U Washington | |
| Jason Moffat | University of Toronto | |
| Eric Brown | McMaster | |
| Phil Hieter | University of British Columbia | |
| Jussi Taipale | Karonlinska Institute | |
| Sarah Jenna | University of Quebec at Montreal | |
| Jens Lagergren | Karolinska University | |
| Mikko Taipale | University of Toronto | |
| Kris Gunsalus | NYU | |
| Michael Boutros | DKFZ | |
| Stirling Churchman | Harvard | |
| Lars Steinmetz | Stanford | |
| Quaid Morris | University of Toronto | |
| Rich Bonneau | NYU | |
| Sebastian Nijmen | Oxford U | |
| Thijn Brummelkamp | Netherland Cancer Institute | |
| Wolfgang Huber | EMBL | |
| Gregor Jansen | McGill University | |
| Bjorn Fjukstad | The Arctic University of Norway |
The focus of this meeting was on computational models of tumorigenesis and tumoral evolution, and studies of early breast lesions.
 |
This meeting brought down many members of the international microRNA community wheree we discussed research, current challenges and the way onward.
 |
| Organizers | Institution | |
| Thomas Duchaine | McGill University, Canada | |
| Vanessa Dumeaux | McGill University, Canada | |
| Mike Hallett | McGill University, Canada | |
| Invitees | Institution | |
| Julie Pelloux | Universite de Montreal | |
| Chris Hillier | University of the West Indies | |
| Ali Tofigh | McGill University | |
| Frank Slack | Yale University | MicroRNA-based therapeutics in cancer |
| Brenda Andrews | University of Toronto | Using yeast functional genomics to explore biological pathways and non-coding RNA function |
| Eric Lai | Memorial Sloan-Kettering Cancer Center | MicroRNAs and other post-transcriptional regulation |
| Jens Lagergren | SciLifeLab Stockholm | |
| Marie Öhman | Stockholm University | Regulation of miRNAs by RNA editing in neurons |
| Chris Hammell | Cold Spring Harbor Laboratory | Temporal regulation of miRNA expression during C. elegans post-embryonic development |
| René Ketting | Institute of Molecular Biology (IMB) | Diversity of small RNA pathways in germ cells |
| Colin Crist | McGill University | Post-transcriptional mechanisms regulating skeletal muscle stem cell activity |
| Martin Simard | Laval University Cancer Research Center | Understanding small RNA pathways through the Argonautes |
| Marc Fabian | Lady Davis Institute for Medical Research | Insights into recruitment of the CCR4-NOT complex by mammalian gene silencing platforms |
| Amy Pasquinelli | UC San Diego | The multifaceted miRISC |
| Helge Grosshans | Friedrich Miescher Institute for Biomedical Research | Towards a quantitative understanding of miRNA function and regulation in physiology |
| Antonio Giraldez | Yale University | Using ribosome footprinting to analyze the coding genome |
| Eric Miska | Gurdon Institute | C. elegans piRNAs |
| Nada Jabado | McGill University | Fusion of TTYH1 with the C19MC microRNA cluster in the embryonal brain tumor ETMR |
| Tim Hughes | University of Toronto | Exploring the universe of RNA-binding proteins |
| Sohail Tavazoie | The Rockefeller University | microRNAs as molecular probes into the biology of metastasis |
| Nikolaus Rajewsky | Max Delbruck Center for Molecular Medicine | RNA-protein interactions |
| Richard Carthew | Northwestern University | MicroRNAs in Drosophila |
| Hin Hark Gan | New York University | 3D Modeling of miRISC-Target Interactions |
| Yifei Yan | Universite de Montreal | Designing small artificial miRNAs to inhibit HIV production |
| Paul Boutros | Ontario Institute for Cancer Research (OICR) | miRNA-Based Biomarkers for Cancer: Challenges & Successes |
| Nathanael Weill | Universite de Montreal | Modeling miRNA-induced gene expression regulation |
| Lill-Tove Busund | Universitetet i Tromsø | Detection of microRNA in tissue blocks or MicroRNA in the clinic |
| Line Moi | University hospital of North Norwayl | microRNAs as biomarkers in breast cancer |
| Francois Major | Universite de Montreal | RNA transient structures control the microRNA maturation pathway |
| Ahilya Sawh | McGill University | The post-translational life of DCR-1 |
| Ariel Donayo | McGill University | Regulation of the maturation of pri-miR-17~92 in normal and cancer cells |
| Mathieu Flamand | McGill University | A non-canonical site reveals multiple cooperative mechanisms in microRNA-mediated silencing |
This meeting focused on the development of genomics-based approaches for the identification and implementation of biomarkerss for patient prognosis and benefit to therapy.
| Organizers | Institution | |
| Mike Hallett | McGill University, Canada | |
| Vanessa Dumeaux | McGill University, Canada | |
| Sylvie Mader | Universite de Montreal, Canada | |
| Invitees | Institution | |
| Justyna Kulpa | Universite de Montreal | Endocrinology |
| Uri David Akavia | Columbia University | Computational Systems Biology |
| David Cotnoir-White | Universite de Montreal | Molecular Oncology |
| Quaid Morris | University of Toronto, Canada | Computational Biology |
| Trevor Levin | OHSU Knight Cancer Institute, Oregon | Cancer Genomics |
| Sandra Krum-Hansen | University of Tromso | Epidemiology |
| Luke McCaffrey | McGill University, Canada | Molecular Oncology |
| Veena Sangwan | University of Minnesota | Molecular Oncology |
| Jennifer Knight | Universite de Montreal | Molecular Oncology |
| Wilbert Zwart | Universite de Montreal | Molecular Oncology |
| Eric Paquet | McGill University, Canada | Computational Biology |
| John Ozcelik | McGill University, Canada | Computational Biology |
| Sadiq Saleh | McGill University, Canada | Cancer Genomics |
| Matthew Suderman | McGill University, Canada | Computational Biology |
| Ali Tofigh | McGill University, Canada | Computational Biology |
| Robert Lesurf | McGill University, Canada | Computational Biology |
| Sean Cory | McGill University, Canada | Computational Biology |
| Julie Livingstone | McGill University, Canada | Computational Biology |
| Harvey Smith | McGill University, Canada | Molecular Oncology |
| Genevieve Deblois | McGill University, Canada | Molecular Oncology |
| Jose Teodoro | McGill University, Canada | Molecular Oncology |
| Lodewyk Wessels | Universite de Montreal | Computational Biology |
| Mathieu Lupien | Universite de Montreal | Molecular Oncology |
| Nicholas Bertos | McGill University, Canada | Molecular Oncology |
| Morag Park | McGill University, Canada | Molecular Oncology |
| Vincent Giguere | McGill University, Canada | Molecular Oncology |
| Thomas Duchaine | McGill University, Canada | Molecular Oncology |
| David Juncker | McGill University, Canada | Microfluidics |
| Therese Sorlie | Radition Hopsital, Oslo | Breast Cancer Genomics |
| Eiliv Lund | Universite de Montreal | Epidemiology |
| Marc Zapatka | Universite de Montreal | Cancer Genomics and Bioinformatics |
| Benjamin Haibe-Kains | Universite de Montreal | Computational Biology |
| Steve Jones | Universite de Montreal | Genomics |
| Janusz Rak | McGill University, Canada | Molecular Oncology |
| Simak Ali | Imperial College School of Medicine, London | Molecular Oncology |
| Josie Dostie | McGill University, Canada | Molecular Oncology |
| Paul Boutros | Ontario Institute for Cancer Research, Canada | Cancer Informatics |
| Stephen M. Robbins | Southern Alberta Cancer Research Institute, Canada | Extracellular signalling in Cancer |
| Richard Simon | National Cancer Institute, USA | Cancer Treatment and Diagnosis |
This meeting focused on transcription factors, protein-DNA binding and the complex network of regulatory dependencies they induce.
| Organizers | Institution | |
| Mike Hallett | McGill University, Canada | |
| Tim Hughes | University of Toronto, Canada | |
| Organizers | Institution | |
| Brenda Andrews | University of Toronto, Canada | Yeast network biology |
| Phil Benfey | Duke University, USA | Gene expression in Arabidopsis |
| Kris Gunsalus | New York University, USA | C. elegans functional genomics and post-transcriptional regulation |
| Henry Krause | University of Toronto, Canada | transcriptional and post-transcriptional regulation in Drosophila and zebrafish |
| Jason Lieb | UNC Chapel Hill, USA | Functional genomics, gene regulation, and chromatin |
| Howard Lipshitz | University of Toronto, Canada | posttranscriptional regulation of gene expression in Drosophila |
| Don Moerman | University of British Columbia, Canada | Basic genetics, transposon biology, genetic methodology, genomics and muscle development in Caenorhabditis elegans |
| Quaid Morris | University of Toronto, Canada | Computational biology |
| Corey Nislow | University of Toronto, Canada | Systems biology |
| Fabio Piano | New York University, USA | Systems biology, C. elegans embryogenesis |
| Eran Segal | Weizmann Institute, Israel | Computational biology |
| Craig Smibert | University of Toronto, Canada | Regulation of translation |
| Jussi Taipale | Karolinska Institutet, Sweden | RNAi screening, identification of transcriptional regulatory mechanisms, and identification of the sequence specificities of DNA-binding proteins |
| Charles Vinson | National Institutes of Health, USA | promoter analysis, transcription factors, DNA methylation |
| Bob Waterston | University of Washington, USA | Genome science |
| Lisa Stubbs | University of Illinois, USA | C2H2 zinc finger transcription factors |
| Nick Luscombe | EBI, UK | Computational biology |
| Alain Nepveu | McGill University, Canada | Transcriptional regulation in mammalian cells |
| Matt Weirauch | University of Toronto, Canada | Computational methods for transcription regulation |
| Anna Lee | University of Toronto, Canada | |
| Kate Cook | University of Toronto, Canada | |
| Hilal Kazan | University of Toronto, Canada | |
| Xiao Li | University of Toronto, Canada | |
| Guillaume Bourque | McGill University and Genome Quebec Innovation Center, Canada | |
| Jens Lagergren | Science for Life Laboratory, Sweden | |
| Susanne Groebner | University of Heidelberg, Germany | |
| Benjamin Boucher | University of Quebec in Montreal, Canada |
The central topic of this meeting was to explore the use of techniques from systems biology including high-throughput profiling, modelling and multivariate biomarker panels in the context of breast cancer.
| Organizers | Institution | |
| Mike Hallett | McGill University, Canada | |
| Vanessa Dumeaux | University of Tromso, Norway | |
| Participants | Affiliation | Expertise |
| Nicholas Bertos | McGill University, Canada | Genomics |
| Margarethe Biong | Oslo University Hospital, Norway | Genomics |
| Sean Cory | McGill University, Canada | Bioinformatics |
| Slim Fourati | Universite de Montreal, Canada | Genomics |
| Gregor Jansen | McGill University, Canada | Cell Signaling |
| Vessela Kristensen | Oslo University Hospital, Norway | Genomics |
| Hege Landmark-Hoyvik | Oslo University Hospital, Norway | Genomics |
| Robert Lesurf | McGill University, Canada | Bioinformatics |
| Julie Livingstone | McGill University, Canada | Bioinformatics |
| Joseph Lucas | Duke University, USA | Bioinformatics |
| Sylvie Mader | Universite de Montreal, Canada | Genomics |
| Hans-Kristian Moen-Vollan | Oslo University Hospital, Norway | Surgeon; Genomics |
| William Muller | McGill University, Canada | Molecular Oncology |
| Morag Park | McGill University, Canada | Molecular Oncology |
| Sandra Caroline Plancade | University of Tromso, Norway | Statistics |
| Irene Pylypenko | McGill University, Canada | Bioinformatics |
| Hege Russness | Harvard University, USA | Breast Cancer |
| Derek Ruths | McGill University, Canada | Bioinformatics |
| Cenk Sahinalp | Simon Fraser University, Canada | Computational Genomics |
| Sadiq Saleh | McGill University, Canada | Bioinformatics |
| Babette Schade | McGill University, Canada | Molecular Oncology |
| Bita Sehat | McGill University, Canada | Molecular Oncology |
| Sohrab Shah | University of British Columbia, Canada | Bioinformatics |
| Peter Siegel | McGill University, Canada | Molecular Oncology |
| Matthew Suderman | McGill University, Canada | Bioinformatics |
| David Thomas | McGill University, Canada | Other |
| Christina Tognon | University of British Columbia, Canada | Breast Cancer |
| Ali Tofigh | McGill University, Canada | Bioinformatics |
| Braaten Tonje | University of Tromso, Norway | Epidemiology |
| Josie Ursini-Siegel | Universite de Montreal, Canada | Molecular Oncology |
| Johan Vallon-Christersson | Lund University, Sweden | BC Susceptibility Genes |
| John White | McGill University, Canada | Genomics |
| Organizers | Institution | |
| Mike Hallett | McGill University | |
| Sarah Jenna | Université de Québec à Montréal | |
| David Thomas | McGill University | |
| Invitees | ||
| Brenda Andrews | University of Toronto | Study of cell cycle and cell division in budding yeast |
| Suzana Anjos | McGill University | Cystic fibrosis, response-to-chemical gene expression signatures |
| Bill Balch | The Scripps Research Institute | Protein trafficking and misfolding disease |
| Margarethe Biong | Norwegian Radium Hospital | Associations between genotype profiles and breast cancer risk factors |
| Rich Bonneau | New York University | Peptide design |
| Fabiana Ciciriello | McGill University | Cystic fibrosis, response-to-chemical gene expression signatures |
| Vanessa Dumeaux | University of Tromso | Epidemiology of Environmental Exposures in Cancer |
| Elias Epp | McGill University | Fungal genomics, antifungal synergy |
| Guri Giaever | University of Toronto | Chemical genomics |
| Sara JC Gosline | McGill University | Probabilistic models of endoplasmic reticulum quality control |
| Gregor Jansen | McGill University | Chemogenomic profiling, antifungal synergy |
| Danielle Kemmer | McGill University | The study of orphan diseases via chemical genomics |
| Hege BK Landmark-Høyvik | Norwegian Radium Hospital | Blood gene expression to investigate response to and late side effects of radiotherapy |
| Anna Y Lee | McGill University | Predicting antifungal synergy and therapeutic targets for genetic disorders |
| Sébastien Lemieux | Université de Montréal | Functional and structural bioinformatics |
| Nicolas Moitessier | McGill University | Chemo-informatics |
| Corey Nislow | University of Toronto | Chemical genomics |
| Dana Pe'er | Columbia University | Computational systems biology |
| Lekha Sleno | Université de Québec à Montréal | Metabolome, drug metabolism and mass spectrometry |
| Jérome Waldispühl | McGill University | Predicting the structures of proteins and RNAs |
| Dan Zilberstein | Technion - Israel Institute of Technology | Leishmaniasis |
This meeting focused on genomics approaches in breast cancer to study both murine models and the human disease.
| Organizers | Institution | |
| Mike Hallett | McGill University | |
| Morag Park | McGill University | |
| Invitees | ||
| Jørgen Aarøe | Rikshospitalet Institute for Cancer Research | Genomic alterations and systemic changes in early breast cancer. |
| Nicholas Bertos | McGill University | DTCs/CTCs in breast cancer. |
| Sean Cory | McGill University | Bioinformatics of breast cancer. |
| Rachelle Dillon | McGill University | Erbb2 mouse models of breast cancer. |
| Vanessa Dumeaux | Rikshospitalet Institute for Cancer Research/Princeton | Breast density, epidemiology. |
| Slim Fourati | University of Montreal | Estrogen positive breast cancer. |
| Kalle Gehring | McGill University | Structural biology, endocytosis. |
| Vincent Giguère | McGill University | Receptor signaling in breast cancer. |
| Kun Huang | Ohio State University | Bioinformatics of cancer. |
| Gregor Jansen | McGill University | Cell signaling pathways. |
| Jennifer Knight | McGill University | Met mouse models of breast cancer. |
| Julie Laferrière | McGill University | Metastases and the microenvironment in breast cancer. |
| Gustavo Leone | Ohio State University | Breast tumor microenvironment. |
| Robert Lesurf | McGill University | Bionformatics in breast cancer. |
| Jason Madore | University of Montreal | Tissue microarrays for ovarian cancer. |
| Anne-Marie Mes-Masson | University of Montreal | Identification and characterization of expression patterns in cancer progression. |
| Aslaug Aamodt Muggerud | Rikshospitalet Institute for Cancer Research | DCIS in breast cancer. |
| William Muller | McGill University | Role of the Epidermal Growth Factor Receptor (EGFR) family in the induction and progression of breast cancer. |
| Alain Nepveu | McGill University | CDP/Cux. |
| Michael Ostrowski | Ohio State University | Breast tumor microenvironment. |
| Jeff Parvin | Ohio State University | The biology of BRCA1. |
| François Pepin | McGill University | Microenvironment in breast cancer. |
| Jill Ranger | McGill University | STAT3. |
| Maya Saleh | McGill University | Apoptosis, inflammation and host response to infections. |
| Babette Schade | McGill University | Mouse models of meastases. |
| Solmaz Shahahlizadeh | McGill University | Metapredictors in breast cancer. |
| Peter Siegel | McGill University | Breast cancer metastases to bone and soft tissues. |
| Sebastien Tabaries | McGill University | Breast cancer metastases. |
| David Thomas | McGill University | Cell signaling pathways and their role in infectious diseases; molecular chaperone systems in the endoplasmic reticulum. |
| Josie Ursini-Siegel | McGill University | Erbb2 mouse models of breast cancer. |
| Charles Vadnais | McGill University | ChIP-Chip/transcription in breast cancer. |
The invitees discusssed protein-protein, genetic and chemical-genetic interactions in both model organsisms and humans.
| Organizers | Institution | |
| Mike Hallett | McGill University | |
| Sarah Jenna | Université de Québec à Montréal | |
| Invitees | ||
| Brenda Andrews | Donnelly CCBR, University of Toronto | Yeast functional genomics, cell cycle |
| Richard Bonneau | Biology/Computer Science, NYU | Regulatory network inference, de novo structure prediction |
| Charlie Boone | BBDMR, Donnelly CCBR, University of Toronto | Yeast genetic interactions, chemical genomics |
| Benedikt Brors | Theoretical Bioinformatics, German Cancer Research Center (DKFZ) | Cancer biology, gene expression analysis |
| Jenny Bryan | Statistics, University of British Columbia | Statistical genomics |
| Susan Celniker | Lawrence Berkeley National Laboratory | Transcription networks and protein complexes in Drosophila |
| Xavier de Bolle | Molecular Biology, University of Namur FUNDP | Brucella melitensis virulence, inter-species interactions |
| Denis Dupuy | CCSB, DCFI, Harvard Medical School | C. elegans promoterome |
| Andrew Fraser | Wellcome Trust Sanger Institute | Signal transduction, genetic interactions in C. elegans |
| Guri Giaever | Donnelly CCBR, University of Toronto | Chemical genomics |
| Sara Gosline | MCB, McGill University | Integrative bioinformatics for ER quality control studies, proteomics |
| Kris Gunsalus | Biology, New York University | Integrating and mining functional genomics data, C. elegans |
| Tim Hughes | BBDMR, Donnelly CCBR, University of Toronto | Functional genomic units, DNA-binding, RNA processing in yeast and mouse |
| Gregor Jansen | Biochemistry, McGill University | Chemical genomics, cell signaling, endoplasmic reticulum |
| Anna Lee | MCB, McGill University | Integrative bioinformatics for chemical genomics, C. elegans genetic interactions |
| Edward Marcotte | CSSB, ICMB, University of Texas | Protein function and interaction informatics |
| Stephen Michnick | Biochemistry, Université de Montréal | Biochemical and genetic networks, HTP protein interaction studies |
| Corey Nislow | BBDMR, Donnelly CCBR, University of Toronto | Nucleosome occupancy, chemical genomics |
| Tony Pawson | SLRI, University of Toronto | Signal transduction |
| Fabio Piano | Biology, NYU | Functional genomics, early embryogenesis in nematodes |
| Jérôme Reboul | Functional Genomics, Marseilles Cancer Research Center, INSERM | Molecular networks implicated in epithelial polarity, C. elegans |
| Fritz Roth | CCSB, Biological Chemistry and Molecular Pharmacology, Harvard Medical School | genetic pathways, alternative splicing, integration informatics |
| Igor Stagljar | Donnelly CCBR, University of Toronto | Protein-protein interactions, drug discovery, genome stability |
| David Thomas | Biochemistry, McGill University | Chemical genomics, cell signaling, endoplasmic reticulum |
| Christine Vogel | CSSB, ICMB, University of Texas | Bioinformatics for genome evolution, protein expression |
| AJ Marian Walhout | Molecular Medicine, Gene Function and Expression, UMass Medical School | Transcription regulatory circuits in C. elegans |
| Monique Zetka | Biology, McGill University | Meiosis in C. elegans |
This meeting brought together researchers interested in understanding how genomic instability, and the evolution of neoplastic genomes, affects cancer progression.
 |
| Organizers | Institution | |
| Mike Hallett | McGill University | |
| Jens Lagergren | Stockholm Bioinformatics Centre | |
| Ben Raphael | Univerisity of California, San Diego | |
| Participants | Affiliation | Expertise |
| Ake Borg | Lund Stem Cell Center | Genomic alterations/gene expression in breast tumors |
| Collin Collins | UCSF Cancer Center | Genomic instability |
| Greg Finak | MCB, Biochemistry, McGill University | Breast Cancer Gene Expression |
| William Muller | Molecular Oncology, McGill University | Her2/neu mouse models of breast cancer |
| Alain Nepveu | Biochemistry, Molecular Oncology, McGill University | Transcriptional regulation, cancer |
| Morag Park | Biochemistry, Molecular Oncology, McGill University | Receptor tyrosine kinases, Met, Breast Cancer |
| Francois Pepin | MCB, Biochemistry, McGill University | Breast Cancer Gene Expression |
| Babette Schade | Molecular Oncology, McGill University | Her2/neu mouse models of breast cancer |
| Peter Siegel | Medicine, McGill University | Breast cancer metastasis |
| Cristina Tognon | Child and Family Research Institute, UBC | Breast cancer, fusion proteins |
| Nicholas Bertos | BCFGG, McGill University | Manager, BC functional genomics platform |
| Jacqueline Hall | MOG/MCB, McGill University | Postdoc, Biostatistics |
| April Rose | Medicine, McGill University | Bone Mets, BC |
| Marisa Ponzo | Biochemistry, McGill University | Met mouse models |
| Sean Cory | MCB, McGill University | Bioinformatics |
| Robert Lesurf | MCB, McGill University | Bioinformatics, Mouse Models |
| Indrani Attiganal | MCB, McGill University | Manager, BC Bioinformatics |
This meeting brought together researchers interested in diseases of the secretory pathway with special emphasis on the endoplasmic reticulum
</tr> </table>This was the first of our Barbados meetings to focus on breast cancer.
</tr> </table>| Organizers | Institution | |
| Michael Hallett | Bioinformatics, McGill | Bioinformatics |
| Morag Park | Oncology, Biochem, Medicine, McGill | Breast Cancer |
| Participant | Affiliation | Specialty |
| T. Brown | U. of Toronto | Microrrays |
| J. Bryan | Statistics, UBC | Microrrays |
| H. Chen | Mol. Oncology, McGill | LCM, Microarrays |
| G. Finak | Biochemistry, McGill | Bioinformatics |
| A. Filali | Inst. du cancer, U. de Mont. | Bioinformatics |
| I. Jurisica | U. of Toronto | Bioinfo/Ovarian Cancer </tr> |
| B. Muller | Mol. Oncology, McGill | Her2 Mouse Models |
| F. Pepin | Bioinformatics, McGill | Bioinfo/TGF-Beta |
| S. Petkiewicz | Oncology, Biochem, Medicine, McGill | Met, Breast Cancer </tr> |
| B. Schade | Mol. Oncology, McGill | Her2 Mouse Models |
| P. Siegel | Medicine, McGill | TGF-Beta/Neu |
| D. Thomas | Biochemistry, McGill | Chemical Genomic Screens |
| C. Tognon | BCRI, UBC | Fusion Proteins |
| Z. Yakhini | Agilent | Microarrays |
| J. Wulfkuhle | Center for Cancer Research, NCI | Proteomics/Breast Cancer |
Our third Barbados workshop focused on proteomics including protein-protein interactions, mass spectrometry and general structural biology.
Our second Barbados workshop focused on the development of algorithms for identifying transcription factor binding sites.
| Organizers | Institution | |||
| Michael Hallett | McGill University, Canada | Mathieu Blanchette | McGill University | |
| Jens Lagergren | Stockholm Bioinformatics Centre, Sweden | David Bryant | McGill University | |
| Harmen Bussemaker | Columbia | |||
| Alessandro Panconesi | La Sapienza di Roma, Italy | |||
| Tzachi Pilpel | Harvard | |||
| Gary Stormo | Washington University | |||
| Martin Tompa | University of Washington | |||
| Wyeth Wasserman | UBC, Canada | |||
| Kaleigh Smith | McGill University | |||
| Lee Ann McCue | Wadsworth Centre, NYS Dept. of Health | |||
| Markus Jalsensius | McGill University |
Our first Barbados workshop focused on the development of algorithms for exploring genome evolution.
| Organizers | Institution | |||
| Michael Hallett | McGill University, Canada | Benny Chor | Technicon, Israel | |
| Jens Lagergren | Stockholm Bioinformatics Centre, Sweden |
|
Tandy Warnow | University of Texas at Austin, U.S.A. |
| Todd Wareham | Memorial University of Newfoundland, Canada | |||
| Pavel Pevzner | University of Southern California, U.S.A. | |||
| Alberto Caprara | Universitš di Bologna, Italy | |||
| Allessandro Panconesi | Universitš La Sapienza di Roma, Italy | |||
| Nadia El-Mabrouk | Universite de Montreal, Canada | |||
| David Bryant | McGill University, Canada | |||
| Bernard Moret | University of New Mexico, U.S.A. | |||
| Louigi Addario-Berry | McGill University, Canada |